Not that I understand the paper, but excerpt follows below.
http://www.sciencedirect.com/science/article/pii/S0002929711004885
In structure-like analyses, membership in multiple ancestry components can be interpreted as admixture, shared ancestry, or even unresolved ancestry. [25] and [69] However, some heuristic interpretations of the ancestry proportions palette in terms of past migrations seem too obvious to be ignored. For example, it was first suggested by the German orientalist Max Müller that ca. 3,500 years ago a dramatic migration of Indo-European speakers from Central Asia (the putative Indo Aryan migration) played a key role in shaping contemporary South Asian populations and was responsible for the introduction of the Indo-European language family and the caste system in India. A few studies on mtDNA and Y-chromosome variation have interpreted their results in favor of the hypothesis, [70], [71] and [72] whereas others have found no genetic evidence to support it. [3], [6], [73] and [74] However, any nonmarginal migration from Central Asia to South Asia should have also introduced readily apparent signals of East Asian ancestry into India (see Figure 2B). Because this ancestry component is absent from the region, we have to conclude that if such a dispersal event nevertheless took place, it occurred before the East Asian ancestry component reached Central Asia. The demographic history of Central Asia is, however, complex, and although it has been shown that demic diffusion coupled with influx of Turkic speakers during historical times has shaped the genetic makeup of Uzbeks75 (see also the double share of k7 yellow component in Uzbeks as compared to Turkmens and Tajiks in Figure 2B), it is not clear what was the extent of East Asian ancestry in Central Asian populations prior to these events. Another example of an heuristic interpretation appears when we look at the two blue ancestry components (Figure 2B) that explain most of the genetic diversity observed in West Eurasian populations (at K = 8), we see that only the k4 dark blue component is present in India and northern Pakistani populations, whereas, in contrast, the k3 light blue component dominates in southern Pakistan and Iran. This patterning suggests additional complexity of gene flow between geographically adjacent populations because it would be difficult to explain the western ancestry component in Indian populations by simple and recent admixture from the Middle East.
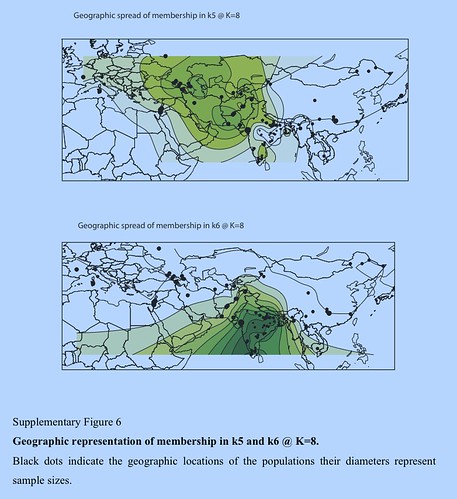
Several aspects of the nature of continuity and discontinuity of the genetic landscape of South Asia and West Eurasia still elude our understanding. Whereas the maternal gene pool of South Asia is dominated by autochthonous lineages, Y chromosome variants of the R1a clade are spread from India (ca 50%) to eastern Europe and their precise origin in space or time is still not well understood.[76]In our analysis we find genetic ancestry signals in the autosomal genes with somewhat similar spread patterns. Both PC2 and k5 light green at K = 8 extend from South Asia to Central Asia and the Caucasus (but not into eastern Europe). In an attempt to explore diversity gradients within this signal, we investigated the haplotypic diversity associated with the ancestry components revealed by ADMIXTURE. Our simulations show that one can detect differences in haplotype diversity for a migration event that occurred 500 generations ago, but chances to distinguish signals for older events will apparently decrease with increasing age because of recombination. In terms of human population history, our oldest simulated migration event occurred roughly 12,500 years ago and predates or coincides with the initial Neolithic expansion in the Near East. Knowing whether signals associated with the initial peopling of Eurasia fall within our detection limits requires additional extensive simulations, but our current results indicate that the often debated episode of South Asian prehistory, the putative Indo-Aryan migration 3,500 years ago (see e.g., Abdulla15) falls well within the limits of our haplotype-based approach. We found no regional diversity differences associated with k5 at K = 8. Thus, regardless of where this component was from (the Caucasus, Near East, Indus Valley, or Central Asia), its spread to other regions must have occurred well before our detection limits at 12,500 years. Accordingly, the introduction of k5 to South Asia cannot be explained by recent gene flow, such as the hypothetical Indo-Aryan migration. The admixture of the k5 and k6 components within India, however, could have happened more recently—our haplotype diversity estimates are not informative about the timing of local admixture.
http://www.sciencedirect.com/science/article/pii/S0002929711004885
Shared and Unique Components of Human Population Structure and Genome-Wide Signals of Positive Selection in South Asia Metspalu, Mait; Romero, Irene�Gallego; Yunusbayev, Bayazit; Chaubey, Gyaneshwer; Mallick, Chandana�Basu; Hudjashov, Georgi; Nelis, Mari; M�gi, Reedik; Metspalu, Ene; Remm, Maido; Pitchappan, Ramasamy; Singh, Lalji; Thangaraj, Kumarasamy; Villems, Richard; Kivisild, Toomas American journal of human genetics doi:10.1016/j.ajhg.2011.11.010 (volume 89 issue 6 pp.731 - 744) -------------------------------------------------------------------------
In structure-like analyses, membership in multiple ancestry components can be interpreted as admixture, shared ancestry, or even unresolved ancestry. [25] and [69] However, some heuristic interpretations of the ancestry proportions palette in terms of past migrations seem too obvious to be ignored. For example, it was first suggested by the German orientalist Max Müller that ca. 3,500 years ago a dramatic migration of Indo-European speakers from Central Asia (the putative Indo Aryan migration) played a key role in shaping contemporary South Asian populations and was responsible for the introduction of the Indo-European language family and the caste system in India. A few studies on mtDNA and Y-chromosome variation have interpreted their results in favor of the hypothesis, [70], [71] and [72] whereas others have found no genetic evidence to support it. [3], [6], [73] and [74] However, any nonmarginal migration from Central Asia to South Asia should have also introduced readily apparent signals of East Asian ancestry into India (see Figure 2B). Because this ancestry component is absent from the region, we have to conclude that if such a dispersal event nevertheless took place, it occurred before the East Asian ancestry component reached Central Asia. The demographic history of Central Asia is, however, complex, and although it has been shown that demic diffusion coupled with influx of Turkic speakers during historical times has shaped the genetic makeup of Uzbeks75 (see also the double share of k7 yellow component in Uzbeks as compared to Turkmens and Tajiks in Figure 2B), it is not clear what was the extent of East Asian ancestry in Central Asian populations prior to these events. Another example of an heuristic interpretation appears when we look at the two blue ancestry components (Figure 2B) that explain most of the genetic diversity observed in West Eurasian populations (at K = 8), we see that only the k4 dark blue component is present in India and northern Pakistani populations, whereas, in contrast, the k3 light blue component dominates in southern Pakistan and Iran. This patterning suggests additional complexity of gene flow between geographically adjacent populations because it would be difficult to explain the western ancestry component in Indian populations by simple and recent admixture from the Middle East.
Several aspects of the nature of continuity and discontinuity of the genetic landscape of South Asia and West Eurasia still elude our understanding. Whereas the maternal gene pool of South Asia is dominated by autochthonous lineages, Y chromosome variants of the R1a clade are spread from India (ca 50%) to eastern Europe and their precise origin in space or time is still not well understood.[76]In our analysis we find genetic ancestry signals in the autosomal genes with somewhat similar spread patterns. Both PC2 and k5 light green at K = 8 extend from South Asia to Central Asia and the Caucasus (but not into eastern Europe). In an attempt to explore diversity gradients within this signal, we investigated the haplotypic diversity associated with the ancestry components revealed by ADMIXTURE. Our simulations show that one can detect differences in haplotype diversity for a migration event that occurred 500 generations ago, but chances to distinguish signals for older events will apparently decrease with increasing age because of recombination. In terms of human population history, our oldest simulated migration event occurred roughly 12,500 years ago and predates or coincides with the initial Neolithic expansion in the Near East. Knowing whether signals associated with the initial peopling of Eurasia fall within our detection limits requires additional extensive simulations, but our current results indicate that the often debated episode of South Asian prehistory, the putative Indo-Aryan migration 3,500 years ago (see e.g., Abdulla15) falls well within the limits of our haplotype-based approach. We found no regional diversity differences associated with k5 at K = 8. Thus, regardless of where this component was from (the Caucasus, Near East, Indus Valley, or Central Asia), its spread to other regions must have occurred well before our detection limits at 12,500 years. Accordingly, the introduction of k5 to South Asia cannot be explained by recent gene flow, such as the hypothetical Indo-Aryan migration. The admixture of the k5 and k6 components within India, however, could have happened more recently—our haplotype diversity estimates are not informative about the timing of local admixture.